1. Harris PC, Torres VE. Polycystic kidney disease, autosomal dominant. In: Adam MP, Ardinger HH, Pagon RA, et al., eds. GeneReviews
® [Internet]. Seattle (WA): University of Washington, 1993-2020 [cited Dec 25, 2021]. Available from:
https://www.ncbi.nlm.nih.gov/books/NBK1246/.
2. Boucher C, Sandford R. Autosomal dominant polycystic kidney disease (ADPKD, MIM 173900, PKD1 and PKD2 genes, protein products known as polycystin-1 and polycystin-2).
Eur J Hum Genet 2004;12:347–354.


3. Rangan GK, Tchan MC, Tong A, Wong AT, Nankivell BJ. Recent advances in autosomal-dominant polycystic kidney disease.
Intern Med J 2016;46:883–892.


4. Perumareddi P, Trelka DP. Autosomal dominant polycystic kidney disease.
Prim Care 2020;47:673–689.


5. Ong AC, Harris PC. Molecular pathogenesis of ADPKD: the polycystin complex gets complex.
Kidney Int 2005;67:1234–1247.


6. Eisenberger T, Decker C, Hiersche M, et al. An efficient and comprehensive strategy for genetic diagnostics of polycystic kidney disease.
PLoS One 2015;10:e0116680.



7. Mantovani V, Bin S, Graziano C, et al. Gene panel analysis in a large cohort of patients with autosomal dominant polycystic kidney disease allows the identification of 80 potentially causative novel variants and the characterization of a complex genetic architecture in a subset of families.
Front Genet 2020;11:464.



8. Kim H, Kim HH, Chang CL, Song SH, Kim N. Novel PKD1 mutations in patients with autosomal dominant polycystic kidney disease.
Lab Med 2021;52:174–180.


9. Nielsen ML, Lildballe DL, Rasmussen M, Bojesen A, Birn H, Sunde L. Clinical genetic diagnostics in Danish autosomal dominant polycystic kidney disease patients reveal possible founder variants.
Eur J Med Genet 2021;64:104183.


10. Pei Y, Obaji J, Dupuis A, et al. Unified criteria for ultrasonographic diagnosis of ADPKD.
J Am Soc Nephrol 2009;20:205–212.



11. Bevilacqua MU, Hague CJ, Romann A, et al. CT of kidney volume in autosomal dominant polycystic kidney disease: accuracy, reproducibility, and radiation dose.
Radiology 2019;291:660–667.


12. Lavu S, Vaughan LE, Senum SR, et al. The value of genotypic and imaging information to predict functional and structural outcomes in ADPKD.
JCI Insight 2020;5:e138724.



13. Chapman AB, Bost JE, Torres VE, et al. Kidney volume and functional outcomes in autosomal dominant polycystic kidney disease.
Clin J Am Soc Nephrol 2012;7:479–486.



14. Ekinci I, Erkoc R, Gursu M, et al. Effects of fasting during the month of Ramadan on renal function in patients with autosomal dominant polycystic kidney disease.
Clin Nephrol 2018;89:103–112.


15. He J, Li Q, Fang S, et al. PKD1 mono-allelic knockout is sufficient to trigger renal cystogenesis in a mini-pig model.
Int J Biol Sci 2015;11:361–369.



16. Kaleta B. The role of osteopontin in kidney diseases.
Inflamm Res 2019;68:93–102.


17. Denhardt DT, Guo X. Osteopontin: a protein with diverse functions.
FASEB J 1993;7:1475–1482.


18. Giachelli CM, Steitz S. Osteopontin: a versatile regulator of inflammation and biomineralization.
Matrix Biol 2000;19:615–622.


21. Kim H, Sung J, Kim H, et al. Expression and secretion of CXCL12 are enhanced in autosomal dominant polycystic kidney disease.
BMB Rep 2019;52:463–468.



22. Irazabal MV, Rangel LJ, Bergstralh EJ, et al. Imaging classification of autosomal dominant polycystic kidney disease: a simple model for selecting patients for clinical trials.
J Am Soc Nephrol 2015;26:160–172.


23. Grantham JJ, Torres VE, Chapman AB, et al. Volume progression in polycystic kidney disease.
N Engl J Med 2006;354:2122–2130.


24. Bhutani H, Smith V, Rahbari-Oskoui F, et al. A comparison of ultrasound and magnetic resonance imaging shows that kidney length predicts chronic kidney disease in autosomal dominant polycystic kidney disease.
Kidney Int 2015;88:146–151.



25. Cornec-Le Gall E, Audrézet MP, Chen JM, et al. Type of PKD1 mutation influences renal outcome in ADPKD.
J Am Soc Nephrol 2013;24:1006–1013.



26. Cornec-Le Gall E, Audrézet MP, Rousseau A, et al. The PROPKD Score: a new algorithm to predict renal survival in autosomal dominant polycystic kidney disease.
J Am Soc Nephrol 2016;27:942–951.


27. Messchendorp AL, Meijer E, Visser FW, et al. Rapid progression of autosomal dominant polycystic kidney disease: urinary biomarkers as predictors.
Am J Nephrol 2019;50:375–385.


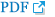
28. Denhardt DT, Noda M, O’Regan AW, Pavlin D, Berman JS. Osteopontin as a means to cope with environmental insults: regulation of inflammation, tissue remodeling, and cell survival.
J Clin Invest 2001;107:1055–1061.



29. Wang H, Sun W, Ma J, Pan Y, Wang L, Zhang W. Polycystin-1 mediates mechanical strain-induced osteoblastic mechanoresponses via potentiation of intracellular calcium and Akt/β-catenin pathway.
PLoS One 2014;9:e91730.



30. Xiao Z, Zhang S, Cao L, Qiu N, David V, Quarles LD. Conditional disruption of Pkd1 in osteoblasts results in osteopenia due to direct impairment of bone formation.
J Biol Chem 2010;285:1177–1187.


31. Xiao Z, Cao L, Liang Y, et al. Osteoblast-specific deletion of Pkd2 leads to low-turnover osteopenia and reduced bone marrow adiposity.
PLoS One 2014;9:e114198.
















 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print