Deep learning outperforms kidney organoid experts
Article information
Over the past decade, the advent of organoid technology has allowed production of laboratory-grown organ tissues as a model system for human biological studies. The use of human organoids has allowed researchers to learn a great deal about human biology and disease mechanisms [1]. Organoids are unique three-dimensional culture systems that are self-organized and similar to actual human organs. Human organoids represent human physiology rather than human-like animal models or two-dimensional culture systems [2].
Kidney organoids model the structure and function of nephrons, which are subdivided into functionally distinct portions, containing podocytes with proximal and distal tubules [3–5]. Kidney organoids have been shown to recapitulate genetic kidney diseases and acute kidney injury, such as cisplatin-induced toxicity [3,4]. Additionally, organoids generated from patient-derived induced pluripotent stem cells (iPSCs) have potential applications in pharmaceutical drug testing and molecular medicine [6]. Recently, protocols for cost-effective bulk production of organoids have been developed and are well-suited for large-scale assays, such as drug screening and regenerative medicine [2,7].
In spite of the availability of kidney organoids, certain technical limitations are associated with the use of organoids. Extended culture of kidney organoids may lead to expansion of nonrenal cell types, including neuronal cells and myofibroblasts [7]. Additionally, patient-derived or genetically modified iPSCs exhibit a variable nature of differentiation and low maturation efficiency. However, these limitations have been partly overcome using strategies to avoid immaturity or screen out unwanted maturation stages.
A study by Park et al. [8] demonstrated that bright-field optical microscopic images could be used to assess the maturity of kidney organoids. They discriminated organoids according to bright-field morphology and examined renal markers of differentiation using quantitative polymerase chain reaction (PCR). These results revealed that bright-field morphology has a high correlation with actual differentiation of organoids. The distinctive feature of this method is that analysis is performed in a living model. The existing methods such as immunofluorescence analysis, reverse transcription-PCR, and single-cell RNA sequencing analysis require destruction of cells in the organoids. Moreover, analysis of bright-field images can be performed in a relatively short time and can be performed at low cost in the laboratory because it is time-efficient and does not use expensive equipment.
Because the analysis of bright-field images is subjective, the outcomes may vary by the observer. To overcome this challenge, the authors adopted deep learning algorithms to objectively analyze bright-field images (Fig. 1). Advances in machine learning, especially deep learning, have allowed exploration, classification, and interpretation of patterns in biological images [9]. One of their typical tasks includes an unsupervised comparison of the features of collections of images by identifying changes in cellular morphology in imaging-based screening [10].
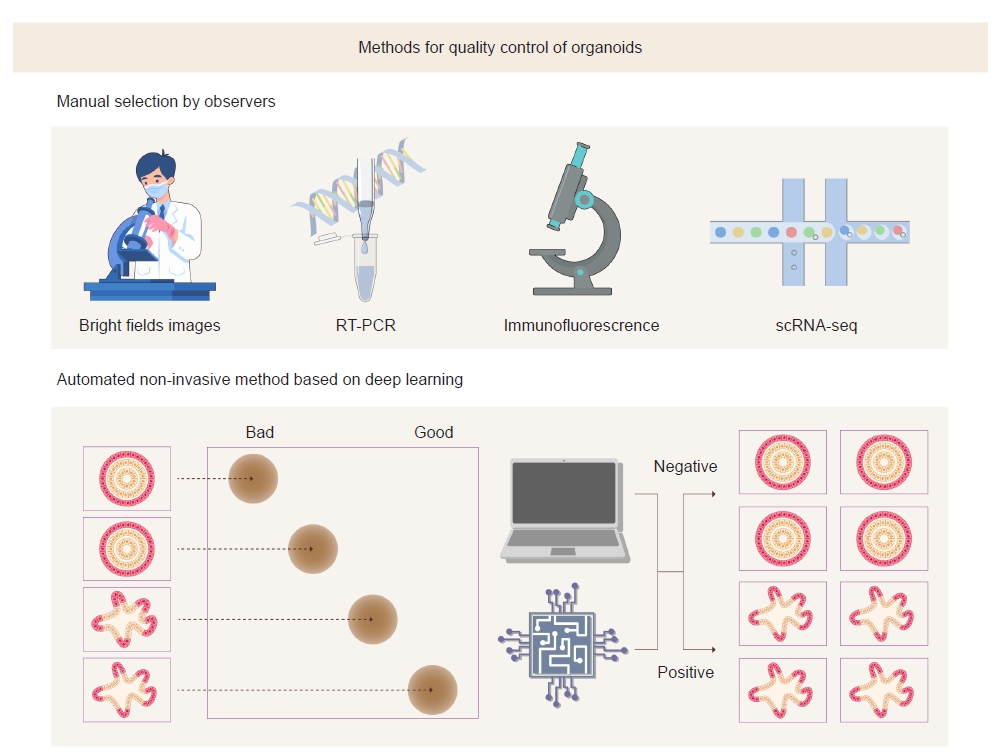
Conventional and deep learning-based methods to evaluate the maturity of kidney organoids.
The deep learning-based method has advantages over conventional methods in the quality control of organoids in that it can reduce subjectivity and variability depending on observers. In addition, bright-field images can be obtained without destruction of organoids.
RT-PCR, reverse transcription polymerase chain reaction; scRNA-seq, single-cell RNA sequencing.
Park et al. [8] trained convolutional neural networks (CNNs) using bright-field images of kidney organoids on day 18 after differentiation and compared the best-performing CNNs with a human-based classifier. They concluded that DenseNet121 is most suitable for predicting the differentiation of kidney organoids. This model classified organoid images into useful (positive) and nonuseful (negative) groups. The results revealed that the CNN algorithm had higher accuracy and speed in classifying organoids than human experts.
Park et al. [8] highlighted that a deep learning model could accurately validate kidney organoid maturity based on analysis of bright-field morphology. The use of bright-field images is a cost-effective and fast strategy for distinguishing suitable organoids. This noninvasive and nondestructive prediction method could also be applied to standardize the quality control protocol of organoids and to high-throughput imaging analysis for drug screening. However, the current model has only been trained on 150 images of “good” and “bad” kidney organoids, which is a relatively small scale. Therefore, its practical use remains challenging. Breakthrough improvements can be achieved by training with many annotated images, which would enable a more detailed classification than just positive or negative groups. Furthermore, using additional learning models and including images for quality control in publicly available datasets would be helpful for improving this tool. We can also expect that this deep learning algorithm is applicable to kidney organoids induced by other differentiation protocols.
Notes
Conflicts of interest
All authors have no conflicts of interest to declare.
Funding
This study was supported by the National Research Foundation of Korea (NRF) funded by the Korean government (MSIT) (2018R1A5A2025079).
Data sharing statement
The data presented in this study are available on request from the corresponding author.
Authors’ contributions
Writing–original draft: SY
Writing–review & editing: HYG
All authors read and approved the final manuscript.
