1. Richards RG, ap Gwynn I. A novel method for viewing heavy metal stained and embedded biological tissue by field emission scanning electron microscopy.
Scanning Microsc 1996;10:111–119.

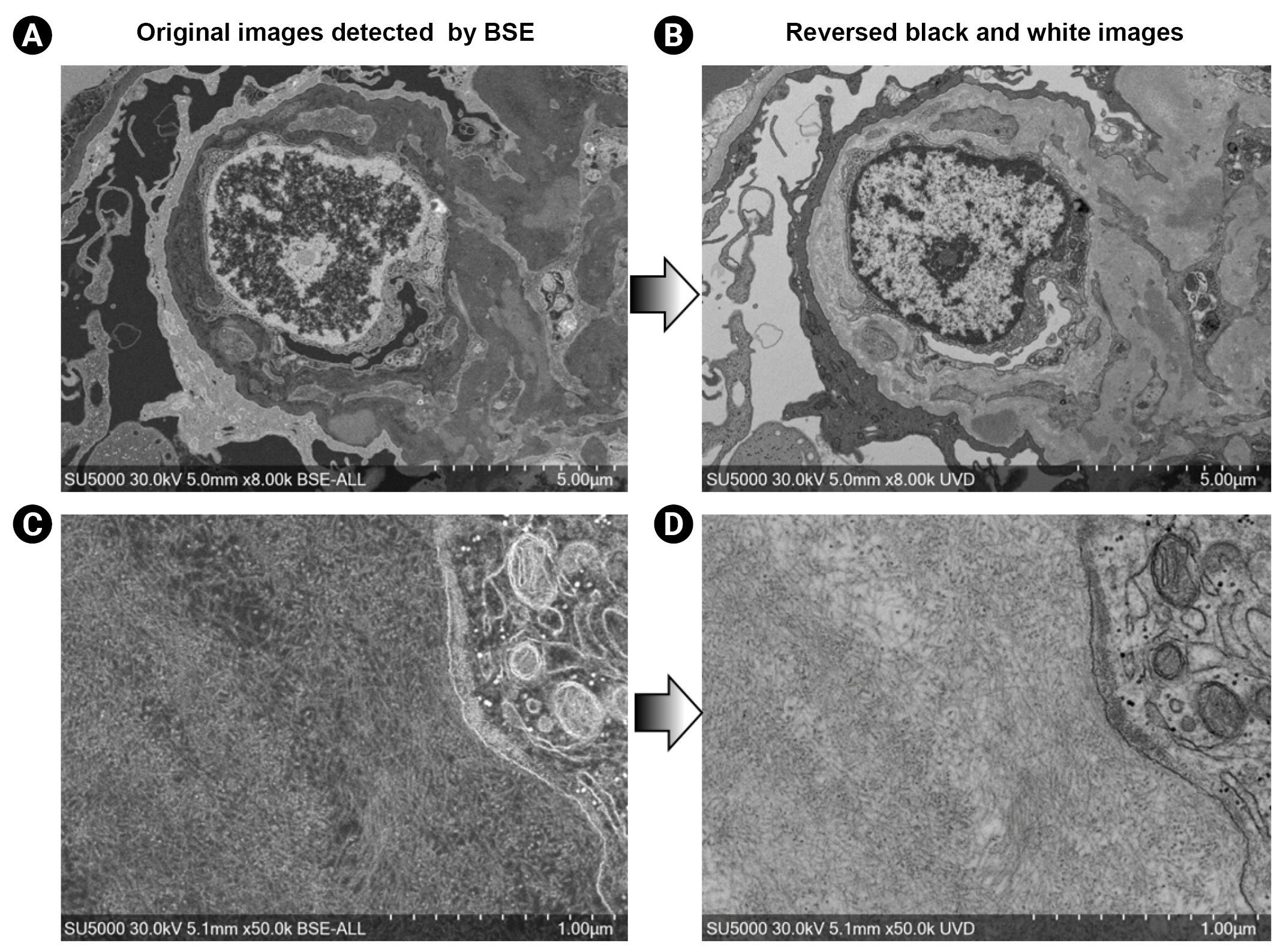
2. Koga D, Kusumi S, Watanabe T. Backscattered electron imaging of resin-embedded sections.
Microscopy (Oxf) 2018;67:196–206.

3. Briggman KL, Bock DD. Volume electron microscopy for neuronal circuit reconstruction.
Curr Opin Neurobiol 2012;22:154–161.


4. Titze B, Genoud C. Volume scanning electron microscopy for imaging biological ultrastructure.
Biol Cell 2016;108:307–323.


5. Kubota Y, Sohn J, Kawaguchi Y. Large volume electron microscopy and neural microcircuit analysis.
Front Neural Circuits 2018;12:98.



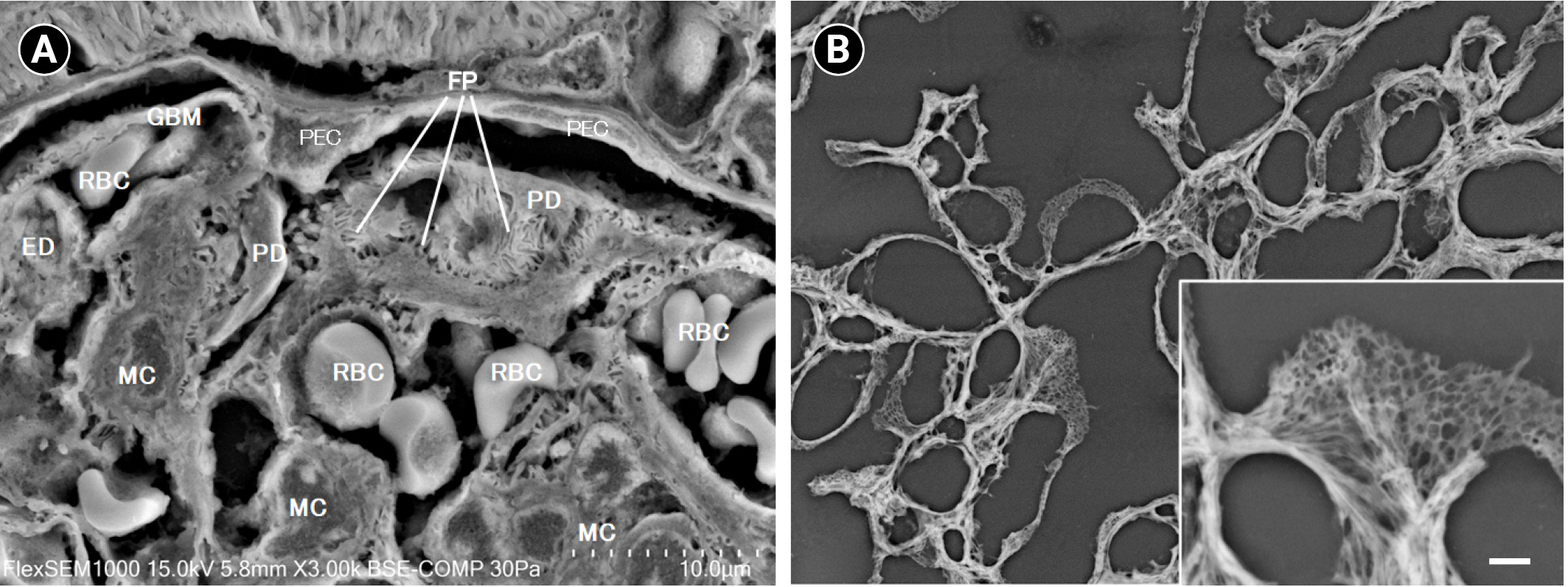
6. Ichimura K, Miyazaki N, Sadayama S, et al. Three-dimensional architecture of podocytes revealed by block-face scanning electron microscopy.
Sci Rep 2015;5:8993.



7. Randles MJ, Collinson S, Starborg T, et al. Three-dimensional electron microscopy reveals the evolution of glomerular barrier injury.
Sci Rep 2016;6:35068.



8. Takaki T, Ohno N, Saitoh S, Nagai M, Joh K. Podocyte penetration of the glomerular basement membrane to contact on the mesangial cell at the lesion of mesangial interposition in lupus nephritis: a three-dimensional analysis by serial block-face scanning electron microscopy.
Clin Exp Nephrol 2019;23:773–781.


9. Nagai M, Saitoh S, Takaki T, et al. Glomerular cellular interactions following disruption of the glomerular basement membrane in IgA nephropathy: ultrastructural analyses by 3-dimensional serial block-face scanning electron microscopy.
Kidney Med 2020;2:222–225.



10. Ichimura K, Kakuta S, Kawasaki Y, et al. Morphological process of podocyte development revealed by block-face scanning electron microscopy.
J Cell Sci 2017;130:132–142.


11. Ichimura K, Miyaki T, Kawasaki Y, Kinoshita M, Kakuta S, Sakai T. Morphological processes of foot process effacement in puromycin aminonucleoside nephrosis revealed by FIB/SEM tomography.
J Am Soc Nephrol 2019;30:96–108.


12. Miyaki T, Kawasaki Y, Hosoyamada Y, et al. Three-dimensional imaging of podocyte ultrastructure using FE-SEM and FIB-SEM tomography.
Cell Tissue Res 2020;379:245–254.


13. Micheva KD, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits.
Neuron 2007;55:25–36.



14. Koike T, Kataoka Y, Maeda M, et al. A device for ribbon collection for array tomography with scanning electron microscopy.
Acta Histochem Cytochem 2017;50:135–140.



15. Kubota Y, Sohn J, Hatada S, et al. A carbon nanotube tape for serial-section electron microscopy of brain ultrastructure.
Nat Commun 2018;9:437.



16. Wacker I, Schroeder RR. Array tomography.
J Microsc 2013;252:93–99.


17. Inaga S, Hirashima S, Tanaka K, et al. Low vacuum scanning electron microscopy for paraffin sections utilizing the differential stainability of cells and tissues with platinum blue.
Arch Histol Cytol 2009;72:101–106.


18. Inaga S, Kato M, Hirashima S, et al. Rapid three-dimensional analysis of renal biopsy sections by low vacuum scanning electron microscopy.
Arch Histol Cytol 2010;73:113–125.


19. Miyazaki H, Uozaki H, Tojo A, et al. Application of low-vacuum scanning electron microscopy for renal biopsy specimens.
Pathol Res Pract 2012;208:503–509.


20. Okada S, Inaga S, Kitamoto K, et al. Morphological diagnosis of Alport syndrome and thin basement membrane nephropathy by low vacuum scanning electron microscopy.
Biomed Res 2014;35:345–350.


21. Okada S, Inaga S, Kawaba Y, et al. A novel approach to the histological diagnosis of pediatric nephrotic syndrome by low vacuum scanning electron microscopy.
Biomed Res 2014;35:227–236.


22. Masuda Y, Yamanaka N, Ishikawa A, et al. Glomerular basement membrane injuries in IgA nephropathy evaluated by double immunostaining for α5(IV) and α2(IV) chains of type IV collagen and low-vacuum scanning electron microscopy.
Clin Exp Nephrol 2015;19:427–435.


23. Yokoyama H, Okada S, Yamada Y, et al. Low-vacuum scanning electron microscopy may allow early diagnosis of human renal transplant antibody-mediated rejection.
Biomed Res 2020;41:81–90.


24. Onishi H, Oguchi H, Shinoda K, et al. Pathological analysis of early transplant glomerulopathy in renal allografts using low-vacuum scanning electron microscopy.
Nephron 2020;144(Suppl 1):71–78.


25. Sawaguchi A, Kamimura T, Yamashita A, et al. Informative three-dimensional survey of cell/tissue architectures in thick paraffin sections by simple low-vacuum scanning electron microscopy.
Sci Rep 2018;8:7479.



26. Mukai S, Takaki T, Nagumo T, et al. Three-dimensional electron microscopy for endothelial glycocalyx observation using Alcian blue with silver enhancement.
Med Mol Morphol 2021;54:95–107.


27. Sjollema KA, Schnell U, Kuipers J, Kalicharan R, Giepmans BN. Correlated light microscopy and electron microscopy.
Methods Cell Biol 2012;111:157–173.


28. Gibson KH, Vorkel D, Meissner J, Verbavatz JM. Fluorescing the electron: strategies in correlative experimental design.
Methods Cell Biol 2014;124:23–54.

29. Takizawa T, Robinson JM. FluoroNanogold is a bifunctional immunoprobe for correlative fluorescence and electron microscopy.
J Histochem Cytochem 2000;48:481–486.


30. Koga D, Kusumi S, Shodo R, Dan Y, Ushiki T. High-resolution imaging by scanning electron microscopy of semithin sections in correlation with light microscopy.
Microscopy (Oxf) 2015;64:387–394.


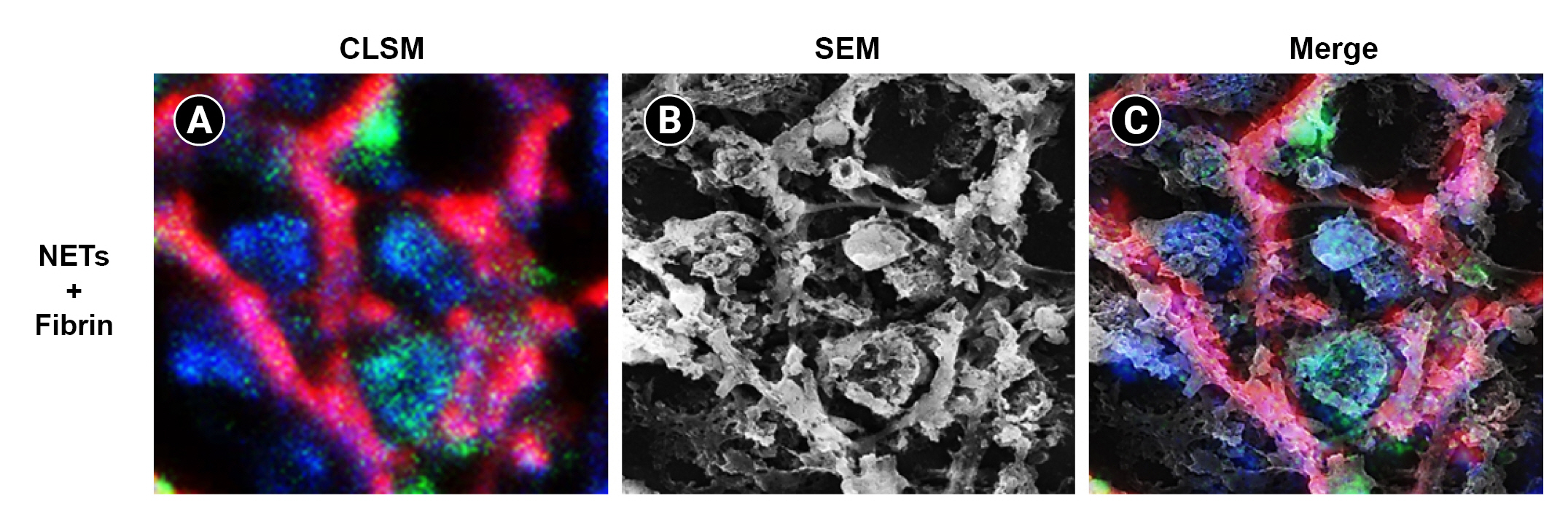
31. Onouchi T, Shiogama K, Mizutani Y, Takaki T, Tsutsumi Y. Visualization of neutrophil extracellular traps and fibrin meshwork in human fibrinopurulent inflammatory lesions: III. Correlative light and electron microscopic study.
Acta Histochem Cytochem 2016;49:141–147.



32. Shibata N, Kohno Y, Findlay SD, Sawada H, Kondo Y, Ikuhara Y. New area detector for atomic-resolution scanning transmission electron microscopy.
J Electron Microsc (Tokyo) 2010;59:473–479.


33. Aoyama K, Takagi T, Hirase A, Miyazawa A. STEM tomography for thick biological specimens.
Ultramicroscopy 2008;109:70–80.


34. Hosoya K, Dan Y, Muto A. Development of STEM imaging in SEM using photon detector.
Microsc Microanal 2019;25(Suppl 2):552–553.











 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print