| Kidney Res Clin Pract > Volume 37(3); 2018 > Article |
|
Abstract
Diabetic kidney disease (DKD) is a major renal complication of diabetes that leads to renal dysfunction and end-stage renal disease (ESRD). Major features of DKD include accumulation of extracellular matrix proteins and glomerular hypertrophy, especially in early stage. Transforming growth factor-β plays key roles in regulation of profibrotic genes and signal transducers such as Akt kinase and MAPK as well as endoplasmic reticulum stress, oxidant stress, and autophagy related to hypertrophy in diabetes. Many drugs targeting the pathogenic signaling in DKD (mostly through protein-coding genes) are under development. However, because of the limited number of protein-coding genes, noncoding RNAs (ncRNAs) including microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) are attracting more attention as potential new drug targets for human diseases. Some miRNAs and lncRNAs regulate each other (by hosting, enhancing transcription from the neighbor, hybridizing each other, and changing chromatin modifications) and create circuits and cascades enhancing the pathogenic signaling in DKD. In this short and focused review, the functional significance of ncRNAs (miRNAs and lncRNAs) in the early stages of DKD and their therapeutic potential are discussed.
The increasing incidence of both type 1 (T1D) and type 2 diabetes (T2D) is a worldwide concern that augments the rates of various micro- and macrovascular complications [1–3]. About 40% of diabetic patients develop diabetic kidney disease (DKD), which leads to chronic kidney disease (CKD) and end-stage renal disease (ESRD) [4–6]. Several biochemical and signal transduction mechanisms leading to DKD have been studied over the years, but the increasing rates of the disease indicate that we need a better understanding of the underlying molecular mechanisms [7–10]. Since the number of protein-coding genes is limited [11,12], noncoding RNAs (ncRNAs) including microRNAs (miRNAs) and long non-coding RNA (lncRNA) have become attractive molecules for identifying drug targets for human diseases. Transcriptome (RNA) sequencing studies have implied that most of the genome could be transcribed into RNA, and many of transcripts are not protein-coding [13,14]. Most of the ncRNAs are lncRNAs (> 200 nucleotides), and there are also small ncRNAs, such as miRNAs [14,15]. LncRNAs have functions in biological processes, gene expression, cell-cycle control, differentiation, and immune responses [16,17]. LncRNAs regulate the expression of genes in cis (neighboring genes) and/or trans (genes genetically far away) locations by several mechanisms including histone-modifying complexes to change chromatin states and modulate the transcription factors or hosting other small RNAs [16–20].
miRNAs are endogenous small ncRNAs (20–25 nucleotides) that usually silence the expression of target genes by hybridizing to the 3′-untranslated regions (3′UTR) of target mRNAs (translational repression and/or mRNA degradation) [21]. Sixty percent of the human protein-coding genes are potentially regulated by miRNAs [21]. miRNAs are expressed as relatively long primary transcripts (pri-miRNAs) transcribed by RNA polymerase II in the nucleus [21–23]. Pri-miRNA is processed to precursor miRNAs (pre-miRNAs, ~70 nucleotide stem-loop structures) by the Drosha/DGCR8 complex in the nucleus. The pre-miRNAs are transported to the cytoplasm by exportin-5 and are cut to mature miRNAs by Dicer in the cytoplasm [21–23]. One strand of mature miRNAs is recruited into the RNA-induced silencing complex (RISC), which contains the Argonaute (Ago) family of proteins that interacts with the 3′UTR of the target mRNAs and induces translational repression or target RNA degradation [21,22]. Although the miRNA targets were predicted by matching seed sequences and flanking sequences by in-silico study [21,24,25], more targets have been identified recently through direct interaction of miRNAs and target RNAs by immunoprecipitation of RISC components (such as Ago) and RNA sequencing strategy [26–28]. These new targets identified by the recent techniques also provide unexpected targets for human diseases. Genetic deletions of Dicer or Drosha in mice cause severe problems in cardiac and renal organs, suggesting the functional relevance of miRNAs in these diseases [29–36].
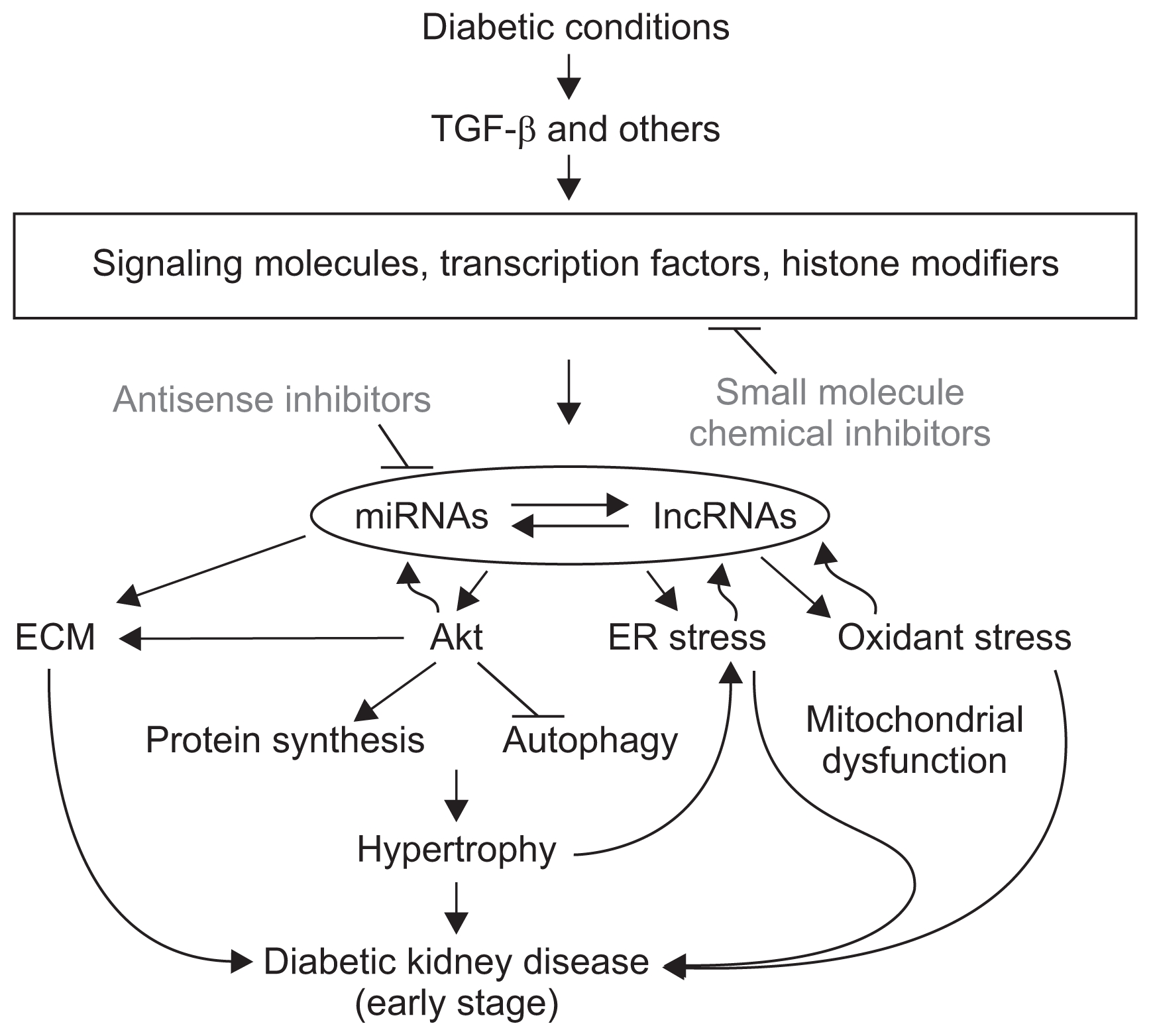
The functions of individual miRNAs provide some hints regarding the mechanistic contribution to DKD progression. Earlier studies on miRNAs in DKD involved miRNAs enriched in kidney or cell-type specific expression in kidney tissues [37,38]. Several miRNAs (miR-192, miR-200b/c, miR-216a, and miR-217) in the kidney are increased in transforming growth factor (TGF)- β1-treated mouse mesangial cells (MMCs) and in renal glomeruli of mouse models of diabetes, streptozotocin (STZ)-injected T1D mice, and T2D db/db mice [39–45]. Those miRNAs play roles in extracellular matrix (ECM) accumulation and hypertrophy (Akt activation) through targeting E-box repressors (ZEB1/2) and PTEN or YBX-1 [39,41,43–45]. Interestingly, some of these miRNAs are creating signal cascades and circuitries to enhance and accelerate the same signals and contribute to the pathogenesis of DKD (Fig. 1). This might be a mechanism of persistent expression of pathologic genes in DKD and other diabetic complications even after controlling hyperglycemia by insulin [1,8,41,43,45–51]. miR-192 is regulated by Smad, p53, and Ets-1 from the transcription factor binding sites in the miR-192 promoter [47,52,53]. The regulation involved epigenetic mechanism through increased histone acetylation by p300 activated by Akt at the miR-192 promoter region, suggesting a link from epigenetics to miRNAs [8,47]. These initial studies have pointed to miR-192 as a master regulator in TGF-β1 response to initiate miRNA cascades and increase expression of ECM genes in DKD [39,41,43–45,47,49,52,53]. miR-192 and miR-215 induce phenotype transition of mesangial cells (MCs) by targeting β-catenin-interacting protein 1 in TGF-β1-treated glomerular MCs and glomeruli from diabetic db/db mice, suggesting multiple functions in DKD [54]. miR-192 expression is also increased in STZ-injected type 1 diabetic mice fed a high-fat diet [42]. Although some reports showed decreased expression of miR-192 in very late stage [55,56], a decrease of this miRNA might be the result, but not the cause of DKD, because miR-192 KO mice did not show any kidney problems and were even protected from DKD [8,51,53]. miR-21 is one of the well-studied miRNAs in DKD, is upregulated in several animal models of DKD and human patients, and targets PTEN, mTOR, matrix metalloproteinases, Smad7, Cdc25a, and Cdk6 [57–60]. More severe renal injuries have been observed in miR-21 KO mice crossed with TGF-β1 transgenic mice [61], and upregulation of miR-21 is protective against renal ischemia–reperfusion injury [62]. miR-93 is a key miRNA (targeting vascular endothelial growth factor-A) that is downregulated in the glomeruli of diabetic db/db mice in renal podocytes and microvascular endothelial cells treated with high glucose condition [63]. Interestingly, miR-93 also targets MSK2, a member of the ribosomal S6 kinase family of serine/threonine kinases, and regulates histone H3 Ser10 phosphorylation (H3S10P) [64,65]. Decreased miR-93 causes an increase in H3S10P and enhances the expression of pathogenic genes through nucleosomal remodeling in DKD [65]. miR-29c was initially identified as an upregulated miRNA under diabetic conditions and activated Rho kinase by targeting Spry-1, related to ECM accumulation and podocyte apoptosis [66]. In cardiac fibrosis, miR-29 targeting collagen was reported [67], and a significant decrease of miR-29 family members and increased collagens were observed in kidney cells from diabetic mice and cells treated with TGF-β1 [68–70]. Interestingly, the anti-diabetic drug linagliptin, a dipeptidyl peptidase-4 (DPP-4) inhibitor, upregulates miR-29 and prevents fibrosis in a mouse model of DKD [71].
Let-7 family members are downregulated in several cancers and also in renal cells treated with TGF-β1, which induces fibrosis through TGF-β1 receptor and collagens [72,73]. Interestingly, the processing of let-7 family members is downregulated by lin28b and upregulated in MMC treated with high glucose or TGF-β1 and in glomeruli from STZ-injected diabetic mice through Smad2/3 activation [74,75]. miR-135a is increased in serum and renal tissue from patients with DKD and db/db mice and promotes ECM accumulation by targeting transient receptor potential cation channel, subfamily C, member 1 (TRPC1) [76]. TGF-β1 decreases miR-130b but upregulates TGF-β1 receptor 1 through unique mechanisms including Ybx1/NFYC targeted by miR-216a, implying another miRNA-mediated amplifying cascade [77]. miR-30 family members are decreased in DKD, and their target connective tissue growth factor (CTGF) is upregulated [78]. miR-22 regulates bone morphogenetic protein-6 (BMP-6) and BMP-7 and further increases TGF-β1 signaling [79]. miR-433 increases TGF-β1 signaling and fibrosis by targeting antizyme inhibitor 1, a regulator of polyamine synthesis through a miRNA-mediated circuit [80]. miR-26a is upregulated by high glucose, activates mTORC1, and enhances hypertrophy and ECM accumulation [81], while the same miRNA is downregulated in DKD patients and also by TGF-β1 in cultured podocytes, leading to an increase of its target CTGF [82]. miR-23b alleviates fibrosis and albuminuria in DKD by targeting Ras GTPase-activating protein SH3 domain-binding protein 2 [83]. miR-146a KO mice showed enhanced proteinuria, renal macrophage infiltration, glomerular hypertrophy, and fibrosis through accelerated inflammation [84] and also showed accelerated glomerular injury and albuminuria in STZ-induced diabetes and podocyte injury through increased ErbB4 and Notch-1, implying that ErbB4/EGFR is a practical target for therapeutics, because several pan-ErbB inhibitors are available [85].
On the other hand, hypertrophy–related miRNAs have been identified. As mentioned above, in the miRNA cascade, miR-216a and miR-217 activate Akt related to hypertrophy by targeting PTEN in the mouse model of DKD and MMC treated with TGF-β1 [41]. The miR-200 family also activates Akt by targeting PI3K inhibitor FOG2 [44]. miR-21 also targets PTEN and activates Akt [86–88]. miR-451 is decreased in early DKD in db/db type 2 diabetic mice and induces hypertrophy through Ywhaz, a protein related to activation of MAPK [89]. miR-214 is also up-regulated by high glucose and induces hypertrophy and matrix expansion by targeting PTEN in renal glomerular mesangial and proximal tubular epithelial cells [90]. miR-NA-181a downregulates DEPTOR (mTOR inhibitor) and activates mTORC2, which activates Akt in TGFβ-induced glomerular MC hypertrophy and ECM protein expression [91]. Autophagy inhibition by the miR-192 cascade induces hypertrophy through Akt activation and FoxO3a phosphorylation [92]. About 40 miRNAs are included in the miR-379 cluster, which is regulated by endoplasmic reticulum (ER) stress in DKD [19]. Because miR-379 targets EDEM3 (inhibitor of ER stress) and miR494 targets Atf3 (repressor of CHOP), upregulation of this miRNA cluster augments ER stress in DKD [19]. ER stress also induces ECM accumulation (fibrosis) and hypertrophy in renal cells (Fig. 1). Interestingly, the miRNA cluster is hosted by an lncRNA (lncMGC, megacluster), which controls expression of the whole cluster [19]. This ER stress-regulated lncMGC is also involved in fibrosis and hypertrophy by regulating the miR-379 miRNA cluster. LncRNA LINC01619 regulates miR-27a/FoxO1 and ER stress-mediated podocyte injury in DKD [93]. LncRNAs (HypERlnc) regulated by hypoxia-induced ER stress are identified in pericytes [94]. Therefore, ER-stress-regulated lncRNAs may have important functions in pathogenesis of kidney diseases. miR-216a and miR-217 were induced by TGF-β1 alone with their host lncRNA, RP23-298H6.1-001 [41].
miR-192 is co-regulated in MCs with its host lncRNA CJ241444, which is induced by TGF-β1 through the promoter Smad binding sites and epigenetic regulation via transcription factor Ets-1 and histone acetylation [47]. Plasmacytoma variant translocation 1 (PVT1), which was identified in a potential locus for diabetic ESRD in a genome-wide single-nucleotide polymorphism genotyping study, increases expression of plasminogen activator inhibitor 1, TGF-β1, and ECM genes in MCs treated with high glucose conditions [95,96]. Five miRNAs (miR-1204, miR-1205, miR-1206, miR-1207, and miR-1208) mapped to the PVT1 locus are also upregulated by high glucose in human MCs and modulate expression of ECM [97,98]. Therefore, those lncRNAs function as host RNAs for small RNAs such as miRNAs [8,51]. LncRNA CYP4B1-PS1-001 is significantly decreased in the early stage in diabetic db/db mouse, and overexpression of the lncRNA inhibits proliferation and fibrosis of MCs [99]. LncRNA MALAT1 (metastasis-associated lung adenocarcinoma transcript) is upregulated in a mouse model of DKD and involved in podocyte injury through β–catenin [100]. Comprehensive screening identified 21 lncRNAs upregulated in two models of renal fibrosis but downregulated in Smad3-knockout mice [101]. Another comprehensive screening of noncoding RNAs by RNA-sequencing has been done in association with early development of DKD [102]. LncRNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy through PGC-1α [103]. LncRNA Erbb4-IR promotes diabetic kidney injury by targeting miR-29b, which regulates collagen genes and ECM accumulation [104]. LncRNA 1700020I14Rik alleviates cell proliferation and fibrosis in DKD through miR-34a-5p/Sirt1/HIF-1α signaling [105]. LncRNA MALAT1 regulates renal tubular epithelial pyroptosis by modulating miR-23c targeting embryonic lethal abnormal vision (ELAVL) 1 in DKD [106]. Those reports also suggest crosstalk between lncRNAs and miRNAs and circuitry mediated by miRNAs and lncRNAs related to fibrotic events, inflammation, and mitochondrial bioenergetics associated with DKD [9,19,41,103,107,108]. Therefore, two major modes of regulation of miRNAs by lncRNAs have been proposed in DKD. First, lncRNAs enhance the expression of miR-NAs by hosting miRNAs or neighboring miRNAs. Second, lncRNAs inhibit the function of miRNAs by hybridizing to miRNAs (sponge function) [109].
Some miRNAs have been implicated in oxidative stress, a major player in DKD pathogenesis. Nox4, a key player in DKD [110–112], was identified as a miR-25 target, suggesting that decreased miR-25 expression upregulates Nox4 to promote oxidative stress and renal dysfunction in rats [113]. Another recent discovery is that the form of MeCP2 phosphorylated by HIPK2, which is known to repress transcription by binding to methylated cytosine, also binds to DGCR8 and inhibits processing of pri-miRNAs [114]. HIPK2 is a key regulator of kidney fibrosis [115], significantly inhibits miR-25 processing (miR-25 reduction and NOX4 increase) through phosphorylation of MeCP2, and was detected in diabetic kidney and MMC treated with TGF-β1. Therefore, the connection from HIPK2 to MeCP2 through miRNA processing is a potential mechanism underlying early stage DKD [116]. Nox4 was also identified as a target of miR-146a, which was down-regulated in high-glucose-treated endothelial cells [117]. miR-205 expression downregulates production of reactive oxygen species through decreases in heme oxygenase and superoxide dismutase (SOD) 1 and 2 [118]. miR-377 was identified as upregulated miRNA by high glucose or TGF-β1 in MCs and increased fibronectin expression and oxidant stress by repression of manganese SOD (Mn-SOD/SOD2) and p21-activated kinase [40]. The miRNA cascade (miR-192, miR-216a, and miR-217) activates Akt and inhibits FoxO3a/SOD2 signaling, leading to oxidative stress in MC [41,119]. Aldose reductase downregulates miR-200a-3p and miR-141-3p, which target Keap1-Nrf2, TGF-β1/TGF-β2, and Zeb1/Zeb2 signaling in MCs and kidneys of diabetic mice [120]. These reports suggest that ncRNAs regulating oxidant stress have critical functions in DKD.
Comprehensive studies of miRNAs in biofluids and tissues provide clues about biomarkers and diagnostics in clinical translational research. Early detection of DKD is extremely useful to prevent progression to renal failure. Several biomarkers, such as peptides, growth factors, and cytokines, of DKD progression have been reported [121]. However, miRNAs are gaining interest as sensitive, noninvasive, and quantitative diagnostic biomarkers for DKD, because of their relatively higher stability in biofluids (urine and plasma and in exosomes) and recent development of detection and quantification methods [122]. Several reports have profiled miRNAs in urine, urinary sediment, urinary exosomes, and in blood or sera of patients with many types of kidney diseases and showed significant correlation with kidney diseases and also with specific stages, fibrosis, renal function (glomerular filtration rate) decline, albuminuria, rapid progression to ESRD, or tissues [8,108,123–138]. LncRNAs can be biomarkers for some human diseases [139]; however, the utility of lncRNAs as biomarkers is not conclusive in DKD. In addition to the above-mentioned publications, more reports showing correlation of ncRNA with kidney diseases and their stages will provide more precise diagnosis of patients by ncRNA profiling.
Many drugs targeting TGF-β or angiotensin II signaling are already under development for treatment of DKD [140,141]. Several reports have shown that miRNAs and lncRNAs are dysregulated in DKD, so targeting them is a potential therapeutic intervention, and some are currently being assessed in trials for preclinical stages [112,129]. miRNA levels can be controlled with miRNA mimics or antisense miRNAs (inhibitor), and stable nuclease-resistant oligonucleotides have been developed for miRNA and lncRNA inhibitors. Locked nucleic acid (LNA) is one of the most potent modifications for inhibiting miRNA activity specifically [41,142,143], and there are LNAs currently in clinical trials [144]. LNA-modified anti-miR-192 specifically and effectively inhibited miR-192, as well as downstream miRNAs (miR-216a, miR-217, and miR-200 family) and p53 in the renal cortex of diabetic mice, and reduced key features of DKD [41,45,53,143]. miR-21 inhibitors also prevented DKD in mouse models [57–60,145]. 2′-O-methyl antisense oligonucleotides targeting miR-29c showed reduced rates of DKD in db/db mice [66]. Targeting lncRNAs is another potential treatment for human diseases [146]. As mentioned above, lncMGC is a hosting ncRNA of the miR-379 cluster. Targeting this lncRNA by LNA-modified antisense oligonucleotides through RNaseH-mediated RNA cleavage (sometimes called GapmeR) [147–149] effectively reduces the expression of lncMGC and cluster miRNAs and attenuates the early features of DKD in a mouse model [19]. MALAT1 siRNA partially restores podocyte function and prohibits β–catenin nuclear accumulation in mouse models of DKD [100]. Therefore, the strategy to inhibit miRNAs and lncRNAs by antisense inhibitors is an encouraging therapeutic approach (Fig. 1).
Some lncRNAs are well-conserved from human to mouse, but other lncRNAs are not. Based on the function of lncRNAs (enhancing miRNA expression or inhibiting miRNA function), even if some lncRNA sequences are not conserved between species, their secondary structures or positions in the genomes are sometimes very similar, and they might have the same functions. In this situation, we need to design species-specific antisense inhibitors against lncRNAs. It might be necessary to redesign human-specific inhibitors to treat patients even if some lncRNA inhibitors work in animal models of DKD.
Optionally, the direct upstream mechanisms controlling the expression/transcription of miRNAs and ln-cRNAs are another therapeutic target (Fig. 1). miR-192 is epigenetically regulated through Ets-1 and histone acetylation, which can be activated by Akt and inhibitors, such as MK-2206, and inhibits miR-192 expression and early features of DKD in mouse models [47]. Several reports have shown Akt activation in animal models of diabetic complication including DKD [119,150–152], and a pharmacological inhibitor of Akt (AS101) showed renoprotection in rat models of diabetes [153]. The anticancer agent paclitaxel decreases miR-192 expression and decreases fibrotic damage in the remnant kidney model [154]. Approaches to upregulate functional miR-NAs are more challenging than inhibition because stable and active molecules are required, although in-vivo delivery methods including adeno-associated virus vectors [129] and bacteriophage MS2 virus-like particles have been tested for overexpressing miRNAs [155]. Targeting negative regulators of miRNAs, such as lin28 (inhibitor of let-7 processing) [72–75] or HIPK2 (inhibitor of miR-25 processing) [114–116,156,157] by siRNA, antisense inhibitors, or chemical inhibitors might be possible (Fig. 1). In fact, small molecule chemical inhibitors of lin28 or HIPK2 (BT173) have been screened and tested to treat cancers or diabetic complications [158–160]. Again, the anti-diabetic drug linagliptin, a DPP-4 inhibitor, upregulates miR-29 and prevents fibrosis in a mouse model of DKD [71].
On the other hand, CRISPR-Cas9 genome editing (and alternatives to Cas9) [161–164] has recently advanced our ability to control genetic and epigenetic changes. The CRISPR-Cas9 system is based on the immune systems in bacteria, which produces degradation in the DNA of invaders such as phages. Using this method in mammalian systems, it is getting easier and faster to introduce site-specific mutations, large deletions, and replacements even in mammalian cells and to create mutant animals. The method can also introduce point mutations or correct mutations in wild-type sequences [165–168]. Another advantage of the system is locus–specific control of gene activation or repression using a protein fusion of Cas9 with transcriptional activators or repressors [169–171]. More interestingly, using a fusion protein of Cas9 with DNA methyltransferase, histone modifying enzymes, or other DNA or histone modifying enzymes, it is possible to change the epigenetic marks, DNA methylation, histone acetylation, or methylation in the locus-specific manner [172–176]. Because epigenetic changes near disease-related genes are critical for DKD and persistent expression of disease genes, site-specific epigenetic modification must be more effective for use in patient treatment since known epigenetic modifying drugs usually have genome-wide (nonspecific) effects, which may cause side effects.
In conclusion, miRNAs and lncRNAs are involved in progression of DKD by targeting genes related to fibrosis, hypertrophy, ER stress, inflammation, oxidant stress, and signal transduction [8,49,112]. An interesting fact is that molecular events in the early stages of disease, such as ECM, hypertrophy, ER stress, and oxidative stress, are frequently observed before later features such as proteinuria in mouse models, and targeting the ncRNAs (early molecules) is effective at reducing the occurrence of such late features [8,19,51,53,143]. Therefore, early detection and treatment of patients before physiological changes such as proteinuria might be necessary to prevent DKD. Because molecular pathogenesis sometimes varies from patient to patient, patient-specific treatments targeting specific ncRNAs (precision medicine) must be considered in treatment of DKD patients [8,70,108]. Some of these RNAs create amplifying cascades and circuits. Therefore, targeting a single miRNA may not be sufficient for DKD treatment. A combination of multiple ncRNAs might be necessary for future treatment of DKD, especially in the early stage, although further study of the expression and functions of those RNAs is necessary.
Figure 1
A proposed model of diabetic kidney disease (DKD) mediated by ncRNAs and potential targets for prevention and treatment
Several ncRNAs (miRNAs and lncRNAs) are involved in the progression of DKD by targeting genes related to ECM accumulation, hypertrophy, ER stress, inflammation, oxidative stress, and signal transduction, especially in the early stages. Some of these key RNAs and signaling molecules create amplifying cascades and circuits that activate each other and accelerate the same signals. The miRNAs and lncRNAs are direct or indirect potential targets for prevention of DKD. Direct upstream factors that control ncRNA expression can also be alternative targets (see main text for details). ECM, extracellular matrix; ER, endoplasmic reticulum; lncRNA, long noncoding RNA; miRNA, micro RNA; ncRNA, noncoding RNA; TGF, transforming growth factor.
References
2. Shah MS, Brownlee M. Molecular and cellular mechanisms of cardiovascular disorders in diabetes. Circ Res 118:1808–1829. 2016;



4. Fineberg D, Jandeleit-Dahm KA, Cooper ME. Diabetic nephropathy: diagnosis and treatment. Nat Rev Endocrinol 9:713–723. 2013;


5. Jones CA, Krolewski AS, Rogus J, Xue JL, Collins A, Warram JH. Epidemic of end-stage renal disease in people with diabetes in the United States population: do we know the cause? Kidney Int 67:1684–1691. 2005;


6. Sarnak MJ, Levey AS, Schoolwerth AC, et al. Kidney disease as a risk factor for development of cardiovascular disease: a statement from the American Heart Association Councils on Kidney in Cardiovascular Disease, High Blood Pressure Research, Clinical Cardiology, and Epidemiology and Prevention. Circulation 108:2154–2169. 2003;


7. Kanwar YS, Sun L, Xie P, Liu FY, Chen S. A glimpse of various pathogenetic mechanisms of diabetic nephropathy. Annu Rev Pathol 6:395–423. 2011;



8. Kato M, Natarajan R. Diabetic nephropathy--emerging epigenetic mechanisms. Nat Rev Nephrol 10:517–530. 2014;



9. Meng XM, Nikolic-Paterson DJ, Lan HY. TGF-β: the master regulator of fibrosis. Nat Rev Nephrol 12:325–338. 2016;


10. Reidy K, Kang HM, Hostetter T, Susztak K. Molecular mechanisms of diabetic kidney disease. J Clin Invest 124:2333–2340. 2014;



11. Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature 409:860–921. 2001;


12. Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science 309:1559–1563. 2005;


13. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. 2012;



14. Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458:223–227. 2009;



15. Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25:1915–1927. 2011;



17. Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol 220:126–139. 2010;


18. Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res 40:6391–6400. 2012;



19. Kato M, Wang M, Chen Z, et al. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat Commun 7:128642016;



20. Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci Adv 3:eaao21102017;



22. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9:102–114. 2008;


23. Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 6:376–385. 2005;


24. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20. 2005;


25. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife 4:e050052015;



26. Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460:479–486. 2009;



27. Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153:654–665. 2013;



28. Van Nostrand EL, Pratt GA, Shishkin AA, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13:508–514. 2016;



29. Harvey SJ, Jarad G, Cunningham J, et al. Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J Am Soc Nephrol 19:2150–2158. 2008;



30. Ho J, Ng KH, Rosen S, Dostal A, Gregory RI, Kreidberg JA. Podocyte-specific loss of functional microRNAs leads to rapid glomerular and tubular injury. J Am Soc Nephrol 19:2069–2075. 2008;



31. Ho J, Pandey P, Schatton T, et al. The pro-apoptotic protein Bim is a microRNA target in kidney progenitors. J Am Soc Nephrol 22:1053–1063. 2011;



32. Shi S, Yu L, Chiu C, et al. Podocyte-selective deletion of dicer induces proteinuria and glomerulosclerosis. J Am Soc Nephrol 19:2159–2169. 2008;



33. Nagalakshmi VK, Ren Q, Pugh MM, et al. Dicer regulates the development of nephrogenic and ureteric compartments in the mammalian kidney. Kidney Int 79:317–330. 2011;


34. Zhdanova O, Srivastava S, Di L, et al. The inducible deletion of drosha and microRNAs in mature podocytes results in a collapsing glomerulopathy. Kidney Int 80:719–730. 2011;



35. Zhao Y, Ransom JF, Li A, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 129:303–317. 2007;


36. Chen JF, Murchison EP, Tang R, et al. Targeted deletion of dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A 105:2111–2116. 2008;



37. Sun Y, Koo S, White N, et al. Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res 32:e1882004;



38. Tian Z, Greene AS, Pietrusz JL, Matus IR, Liang M. MicroR-NA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis. Genome Res 18:404–411. 2008;



39. Kato M, Zhang J, Wang M, et al. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of e-box repressors. Proc Natl Acad Sci U S A 104:3432–3437. 2007;



40. Wang Q, Wang Y, Minto AW, et al. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J 22:4126–4135. 2008;



41. Kato M, Putta S, Wang M, et al. Tgf-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol 11:881–889. 2009;



42. Wang XX, Jiang T, Shen Y, et al. Diabetic nephropathy is accelerated by farnesoid X receptor deficiency and inhibited by farnesoid X receptor activation in a type 1 diabetes model. Diabetes 59:2916–2927. 2010;



43. Kato M, Wang L, Putta S, et al. Post-transcriptional up-regulation of Tsc-22 by Ybx1, a target of miR-216a, mediates TGF-β-induced collagen expression in kidney cells. J Biol Chem 285:34004–34015. 2010;



44. Park JT, Kato M, Yuan H, et al. FOG2 protein down-regulation by transforming growth factor-β1-induced microR-NA-200b/c leads to Akt kinase activation and glomerular mesangial hypertrophy related to diabetic nephropathy. J Biol Chem 288:22469–22480. 2013;



45. Kato M, Arce L, Wang M, et al. A microRNA circuit mediates transforming growth factor-β1 autoregulation in renal glomerular mesangial cells. Kidney Int 80:358–368. 2011;



46. El-Osta A, Brasacchio D, Yao D, et al. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med 205:2409–2417. 2008;



47. Kato M, Dang V, Wang M, et al. TGF-β induces acetylation of chromatin and of Ets-1 to alleviate repression of miR-192 in diabetic nephropathy. Sci Signal 6:ra432013;



48. Cooper ME, El-Osta A, Allen TJ, Watson AMD, Thomas MC, Jandeleit-Dahm KAM. Metabolic karma-the atherogenic legacy of diabetes: the 2017 Edwin Bierman Award Lecture. Diabetes 67:785–790. 2018;


49. Kato M, Natarajan R. MicroRNA circuits in transforming growth factor-β actions and diabetic nephropathy. Semin Nephrol 32:253–260. 2012;



50. Writing Team for the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Research Group. Sustained effect of intensive treatment of type 1 diabetes mellitus on development and progression of diabetic nephropathy: the Epidemiology of Diabetes Interventions and Complications (EDIC) study. JAMA 290:2159–2167. 2003;



51. Kato M, Natarajan R. MicroRNAs in diabetic nephropathy: functions, biomarkers, and therapeutic targets. Ann N Y Acad Sci 1353:72–88. 2015;



52. Chung AC, Huang XR, Meng X, Lan HY. miR-192 mediates TGF-beta/Smad3-driven renal fibrosis. J Am Soc Nephrol 21:1317–1325. 2010;


53. Deshpande SD, Putta S, Wang M, et al. Transforming growth factor-β-induced cross talk between p53 and a microRNA in the pathogenesis of diabetic nephropathy. Diabetes 62:3151–3162. 2013;



54. Mu J, Pang Q, Guo YH, et al. Functional implications of microRNA-215 in TGF-β1-induced phenotypic transition of mesangial cells by targeting CTNNBIP1. PLoS One 8:e586222013;



55. Wang B, Herman-Edelstein M, Koh P, et al. E-cadherin expression is regulated by miR-192/215 by a mechanism that is independent of the profibrotic effects of transforming growth factor-beta. Diabetes 59:1794–1802. 2010;



56. Krupa A, Jenkins R, Luo DD, et al. Loss of microRNA-192 promotes fibrogenesis in diabetic nephropathy. J Am Soc Nephrol 21:438–447. 2010;



57. Wang J, Gao Y, Ma M, et al. Effect of miR-21 on renal fibrosis by regulating MMP-9 and TIMP1 in kk-ay diabetic nephropathy mice. Cell Biochem Biophys 67:537–546. 2013;


58. Zhong X, Chung AC, Chen HY, et al. MiR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia 56:663–674. 2013;


59. Chau BN, Xin C, Hartner J, et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci Transl Med 4:121ra182012;



60. Kölling M, Kaucsar T, Schauerte C, et al. Therapeutic miR-21 silencing ameliorates diabetic kidney disease in mice. Mol Ther 25:165–180. 2017;



61. Lai JY, Luo J, O’Connor C, et al. MicroRNA-21 in glomerular injury. J Am Soc Nephrol 26:805–816. 2015;


62. Xu X, Kriegel AJ, Liu Y, et al. Delayed ischemic preconditioning contributes to renal protection by upregulation of miR-21. Kidney Int 82:1167–1175. 2012;



63. Long J, Wang Y, Wang W, Chang BH, Danesh FR. Identification of microRNA-93 as a novel regulator of vascular endothelial growth factor in hyperglycemic conditions. J Biol Chem 285:23457–23465. 2010;



64. Soloaga A, Thomson S, Wiggin GR, et al. MSK2 and MSK1 mediate the mitogen–and stress–induced phosphorylation of histone H3 and HMG–14. EMBO J 22:2788–2797. 2003;



65. Badal SS, Wang Y, Long J, et al. MiR-93 regulates MSK2-mediated chromatin remodelling in diabetic nephropathy. Nat Commun 7:120762016;



66. Long J, Wang Y, Wang W, Chang BH, Danesh FR. MicroR-NA-29c is a signature microRNA under high glucose conditions that targets sprouty homolog 1, and its in vivo knockdown prevents progression of diabetic nephropathy. J Biol Chem 286:11837–11848. 2011;



67. van Rooij E, Sutherland LB, Thatcher JE, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci 105:13027–13032. 2008;



68. Wang B, Komers R, Carew R, et al. Suppression of microR-NA-29 expression by TGF-β1 promotes collagen expression and renal fibrosis. J Am Soc Nephrol 23:252–265. 2012;



69. Chen HY, Zhong X, Huang XR, et al. MicroRNA-29b inhibits diabetic nephropathy in db/db mice. Mol Ther 22:842–853. 2014;



70. Qin W, Chung AC, Huang XR, et al. TGF-β/Smad3 signaling promotes renal fibrosis by inhibiting miR-29. J Am Soc Nephrol 22:1462–1474. 2011;



71. Kanasaki K, Shi S, Kanasaki M, et al. Linagliptin-mediated DPP-4 inhibition ameliorates kidney fibrosis in streptozotocin-induced diabetic mice by inhibiting endothelial-to-mesenchymal transition in a therapeutic regimen. Diabetes 63:2120–2131. 2014;


72. Brennan EP, Nolan KA, Börgeson E, et al. Lipoxins attenuate renal fibrosis by inducing let-7c and suppressing TGFβR1. J Am Soc Nephrol 24:627–637. 2013;



73. Wang B, Jha JC, Hagiwara S, et al. Transforming growth factor-β1-mediated renal fibrosis is dependent on the regulation of transforming growth factor receptor 1 expression by let-7b. Kidney Int 85:352–361. 2014;


74. Park JT, Kato M, Lanting L, et al. Repression of let-7 by transforming growth factor-β1-induced Lin28 upregulates collagen expression in glomerular mesangial cells under diabetic conditions. Am J Physiol Renal Physiol 307:F1390–F1403. 2014;



75. Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science 320:97–100. 2008;



76. He F, Peng F, Xia X, et al. MiR-135a promotes renal fibrosis in diabetic nephropathy by regulating TRPC1. Diabetologia 57:1726–1736. 2014;


77. Castro NE, Kato M, Park JT, Natarajan R. Transforming growth factor β1 (TGF-β1) enhances expression of profibrotic genes through a novel signaling cascade and microRNAs in renal mesangial cells. J Biol Chem 289:29001–29013. 2014;



78. Wu J, Zheng C, Fan Y, et al. Downregulation of microRNA-30 facilitates podocyte injury and is prevented by glucocorticoids. J Am Soc Nephrol 25:92–104. 2014;


79. Long J, Badal SS, Wang Y, et al. MicroRNA-22 is a master regulator of bone morphogenetic protein-7/6 homeostasis in the kidney. J Biol Chem 288:36202–36214. 2013;



80. Li R, Chung AC, Dong Y, Yang W, Zhong X, Lan HY. The microRNA miR-433 promotes renal fibrosis by amplifying the TGF-β/Smad3-Azin1 pathway. Kidney Int 84:1129–1144. 2013;


81. Dey N, Bera A, Das F, et al. High glucose enhances microRNA-26a to activate mTORC1 for mesangial cell hypertrophy and matrix protein expression. Cell Signal 27:1276–1285. 2015;



82. Koga K, Yokoi H, Mori K, et al. MicroRNA-26a inhibits TGF-β-induced extracellular matrix protein expression in podocytes by targeting CTGF and is downregulated in diabetic nephropathy. Diabetologia 58:2169–2180. 2015;


83. Zhao B, Li H, Liu J, et al. MicroRNA-23b targets Ras GT-Pase-activating protein SH3 domain-binding protein 2 to alleviate fibrosis and albuminuria in diabetic nephropathy. J Am Soc Nephrol 27:2597–2608. 2016;



84. Bhatt K, Lanting LL, Jia Y, et al. Anti-inflammatory role of microRNA-146a in the pathogenesis of diabetic nephropathy. J Am Soc Nephrol 27:2277–2288. 2016;


85. Lee HW, Khan SQ, Khaliqdina S, et al. Absence of miR-146a in podocytes increases risk of diabetic glomerulopathy via up-regulation of ErbB4 and Notch-1. J Biol Chem 292:732–747. 2017;


86. Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133:647–658. 2007;



87. Dey N, Ghosh-Choudhury N, Kasinath BS, Choudhury GG. TGFβ-stimulated microRNA-21 utilizes PTEN to orchestrate AKT/mTORC1 signaling for mesangial cell hypertrophy and matrix expansion. PLoS One 7:e423162012;



88. McClelland AD, Herman-Edelstein M, Komers R, et al. MiR-21 promotes renal fibrosis in diabetic nephropathy by targeting PTEN and SMAD7. Clin Sci (Lond) 129:1237–1249. 2015;


89. Zhang Z, Luo X, Ding S, et al. MicroRNA-451 regulates p38 MAPK signaling by targeting of Ywhaz and suppresses the mesangial hypertrophy in early diabetic nephropathy. FEBS Lett 586:20–26. 2012;


90. Bera A, Das F, Ghosh-Choudhury N, Mariappan MM, Kasinath BS, Ghosh Choudhury G. Reciprocal regulation of miR-214 and PTEN by high glucose regulates renal glomerular mesangial and proximal tubular epithelial cell hypertrophy and matrix expansion. Am J Physiol Cell Physiol 313:C430–C447. 2017;



91. Maity S, Bera A, Ghosh-Choudhury N, Das F, Kasinath BS, Choudhury GG. MicroRNA-181a downregulates deptor for TGFβ-induced glomerular mesangial cell hypertrophy and matrix protein expression. Exp Cell Res 364:5–15. 2018;



92. Deshpande S, Abdollahi M, Wang M, et al. Reduced autophagy by a microRNA-mediated signaling cascade in diabetes-induced renal glomerular hypertrophy. Sci Rep 8:69542018;



93. Bai X, Geng J, Li X, et al. Long noncoding RNA LINC01619 regulates MicroRNA-27a/forkhead box protein O1 and endoplasmic reticulum stress-mediated podocyte injury in diabetic nephropathy. Antioxid Redox Signal 29:355–376. 2018;


94. Bischoff FC, Werner A, John D, et al. Identification and functional characterization of hypoxia-induced endoplasmic reticulum stress regulating lncRNA (HypERlnc) in pericytes. Circ Res 121:368–375. 2017;


95. Alvarez ML, DiStefano JK. Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS One 6:e186712011;



96. Hanson RL, Craig DW, Millis MP, et al. Identification of PVT1 as a candidate gene for end-stage renal disease in type 2 diabetes using a pooling-based genome-wide single nucleotide polymorphism association study. Diabetes 56:975–983. 2007;


97. Huppi K, Volfovsky N, Runfola T, et al. The identification of microRNAs in a genomically unstable region of human chromosome 8q24. Mol Cancer Res 6:212–221. 2008;


98. Alvarez ML, Khosroheidari M, Eddy E, Kiefer J. Role of microRNA 1207-5P and its host gene, the long non-coding RNA Pvt1, as mediators of extracellular matrix accumulation in the kidney: implications for diabetic nephropathy. PLoS One 8:e774682013;



99. Wang M, Wang S, Yao D, Yan Q, Lu W. A novel long non-coding RNA CYP4B1-PS1-001 regulates proliferation and fibrosis in diabetic nephropathy. Mol Cell Endocrinol 426:136–145. 2016;


100. Hu M, Wang R, Li X, et al. LncRNA MALAT1 is dysregulated in diabetic nephropathy and involved in high glucose–induced podocyte injury via its interplay with β–catenin. J Cell Mol Med 21:2732–2747. 2017;



101. Zhou Q, Chung AC, Huang XR, Dong Y, Yu X, Lan HY. Identification of novel long noncoding RNAs associated with TGF-β/Smad3-mediated renal inflammation and fibrosis by RNA sequencing. Am J Pathol 184:409–417. 2014;


102. Tang W, Zhang D, Ma X. RNA-sequencing reveals genome-wide long non-coding RNAs profiling associated with early development of diabetic nephropathy. Oncotarget 8:105832–105847. 2017;



103. Long J, Badal SS, Ye Z, et al. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J Clin Invest 126:4205–4218. 2016;



104. Sun SF, Tang PMK, Feng M, et al. Novel lncRNA Erbb4-IR promotes diabetic kidney injury in db/db mice by targeting miR-29b. Diabetes 67:731–744. 2017;


105. Li A, Peng R, Sun Y, Liu H, Peng H, Zhang Z. LincRNA 1700020I14Rik alleviates cell proliferation and fibrosis in diabetic nephropathy via miR-34a-5p/Sirt1/HIF-1α signaling. Cell Death Dis 9:4612018;



106. Li X, Zeng L, Cao C, et al. Long noncoding RNA MALAT1 regulates renal tubular epithelial pyroptosis by modulated miR-23c targeting of ELAVL1 in diabetic nephropathy. Exp Cell Res 350:327–335. 2017;


107. Lorenzen JM, Thum T. Long noncoding RNAs in kidney and cardiovascular diseases. Nat Rev Nephrol 12:360–373. 2016;


108. Trionfini P, Benigni A, Remuzzi G. MicroRNAs in kidney physiology and disease. Nat Rev Nephrol 11:23–33. 2015;


109. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 17:272–283. 2016;


110. Babelova A, Avaniadi D, Jung O, et al. Role of Nox4 in murine models of kidney disease. Free Radic Biol Med 53:842–853. 2012;


111. Zhu Y, Usui HK, Sharma K. Regulation of transforming growth factor beta in diabetic nephropathy: implications for treatment. Semin Nephrol 27:153–160. 2007;



112. Kato M, Castro NE, Natarajan R. MicroRNAs: potential mediators and biomarkers of diabetic complications. Free Radic Biol Med 64:85–94. 2013;



113. Fu Y, Zhang Y, Wang Z, et al. Regulation of NADPH oxidase activity is associated with miRNA-25-mediated NOX4 expression in experimental diabetic nephropathy. Am J Nephrol 32:581–589. 2010;


114. Cheng TL, Wang Z, Liao Q, et al. MeCP2 suppresses nuclear microRNA processing and dendritic growth by regulating the DGCR8/Drosha complex. Dev Cell 28:547–560. 2014;


115. Jin Y, Ratnam K, Chuang PY, et al. A systems approach identifies HIPK2 as a key regulator of kidney fibrosis. Nat Med 18:580–588. 2012;



116. Oh HJ, Kato M, Deshpande S, et al. Inhibition of the processing of miR-25 by HIPK2-phosphorylated-MeCP2 induces NOX4 in early diabetic nephropathy. Sci Rep 6:387892016;



117. Feng B, Chen S, McArthur K, et al. MiR-146a-mediated extracellular matrix protein production in chronic diabetes complications. Diabetes 60:2975–2984. 2011;



118. Muratsu-Ikeda S, Nangaku M, Ikeda Y, Tanaka T, Wada T, Inagi R. Downregulation of miR-205 modulates cell susceptibility to oxidative and endoplasmic reticulum stresses in renal tubular cells. PLoS One 7:e414622012;



119. Kato M, Yuan H, Xu ZG, et al. Role of the Akt/FoxO3a pathway in TGF-β1-mediated mesangial cell dysfunction: a novel mechanism related to diabetic kidney disease. J Am Soc Nephrol 17:3325–3335. 2006;


120. Wei J, Zhang Y, Luo Y, et al. Aldose reductase regulates miR-200a-3p/141-3pto coordinate Keap1–Nrf2, Tgfβ1/2, and Zeb1/2 signaling in renal mesangial cells and the renal cortex of diabetic mice. Free Radic Biol Med 67:91–102. 2014;


121. Fassett RG, Venuthurupalli SK, Gobe GC, Coombes JS, Cooper MA, Hoy WE. Biomarkers in chronic kidney disease: a review. Kidney Int 80:806–821. 2011;


122. Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A 106:4402–4407. 2009;



123. Szeto CC, Ching-Ha KB, Ka-Bik L, et al. Micro-RNA expression in the urinary sediment of patients with chronic kidney diseases. Dis Markers 33:137–144. 2012;



124. Neal CS, Michael MZ, Pimlott LK, et al. Circulating microR-NA expression is reduced in chronic kidney disease. Nephrol Dial Transplant 26:3794–3802. 2011;


125. Luk CC, Chow KM, Kwok JS, et al. Urinary biomarkers for the prediction of reversibility in acute-on-chronic renal failure. Dis Markers 34:179–185. 2013;



126. Wang G, Kwan BC, Lai FM, Chow KM, Li PK, Szeto CC. Urinary sediment miRNA levels in adult nephrotic syndrome. Clin Chim Acta 418:5–11. 2013;


127. Yang Y, Xiao L, Li J, Kanwar YS, Liu F, Sun L. Urine miR-NAs: potential biomarkers for monitoring progression of early stages of diabetic nephropathy. Med Hypotheses 81:274–278. 2013;



128. Ichii O, Otsuka S, Sasaki N, Namiki Y, Hashimoto Y, Kon Y. Altered expression of microRNA miR-146a correlates with the development of chronic renal inflammation. Kidney Int 81:280–292. 2012;


129. DiStefano JK, Taila M, Alvarez ML. Emerging roles for miRNAs in the development, diagnosis, and treatment of diabetic nephropathy. Curr Diab Rep 13:582–591. 2013;


130. Cai X, Xia Z, Zhang C, et al. Serum microRNAs levels in primary focal segmental glomerulosclerosis. Pediatr Nephrol 28:1797–1801. 2013;



131. Barutta F, Tricarico M, Corbelli A, et al. Urinary exosomal microRNAs in incipient diabetic nephropathy. PLoS One 8:e737982013;



132. Argyropoulos C, Wang K, McClarty S, et al. Urinary microR-NA profiling in the nephropathy of type 1 diabetes. PLoS One 8:e546622013;



133. Higuchi C, Nakatsuka A, Eguchi J, et al. Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metabolism 64:489–497. 2015;


134. Pezzolesi MG, Satake E, McDonnell KP, Major M, Smiles AM, Krolewski AS. Circulating TGF-β1-regulated miRNAs and the risk of rapid progression to ESRD in type 1 diabetes. Diabetes 64:3285–3293. 2015;



135. Baker MA, Davis SJ, Liu P, et al. Tissue-specific microRNA expression patterns in four types of kidney disease. J Am Soc Nephrol 28:2985–2992. 2017;



136. Jia Y, Guan M, Zheng Z, et al. MiRNAs in urine extracellular vesicles as predictors of early-stage diabetic nephropathy. J Diabetes Res 2016:79327652016;



137. Cardenas-Gonzalez M, Srivastava A, Pavkovic M, et al. Identification, confirmation, and replication of novel urinary microRNA biomarkers in lupus nephritis and diabetic nephropathy. Clin Chem 63:1515–1526. 2017;



138. Satake E, Pezzolesi MG, Md Dom ZI, Smiles AM, Niewczas MA, Krolewski AS. Circulating miRNA profiles associated with hyperglycemia in patients with type 1 diabetes. Diabetes 67:1013–1023. 2018;



139. Goyal N, Kesharwani D, Datta M. Lncing non-coding RNAs with metabolism and diabetes: roles of lncRNAs. Cell Mol Life Sci 75:1827–1837. 2018;


140. Declèves AE, Sharma K. New pharmacological treatments for improving renal outcomes in diabetes. Nat Rev Nephrol 6:371–380. 2010;


141. Ruggenenti P, Cravedi P, Remuzzi G. The RAAS in the pathogenesis and treatment of diabetic nephropathy. Nat Rev Nephrol 6:319–330. 2010;


142. Krützfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438:685–689. 2005;


143. Putta S, Lanting L, Sun G, et al. Inhibiting microRNA-192 ameliorates renal fibrosis in diabetic nephropathy. J Am Soc Nephrol 23:458–469. 2012;



144. Lindow M, Kauppinen S. Discovering the first microRNA-targeted drug. J Cell Biol 199:407–412. 2012;



145. Dey N, Das F, Mariappan MM, et al. MicroRNA-21 orchestrates high glucose-induced signals to TOR complex 1, resulting in renal cell pathology in diabetes. J Biol Chem 286:25586–25603. 2011;



146. Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov 12:433–446. 2013;


147. Stein CA, Hansen JB, Lai J, et al. Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res 38:e32010;


148. Michalik KM, You X, Manavski Y, et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 114:1389–1397. 2014;


149. Lee JE, Bennett CF, Cooper TA. RNase H-mediated degradation of toxic RNA in myotonic dystrophy type 1. Proc Natl Acad Sci 109:4221–4226. 2012;



150. Mahimainathan L, Das F, Venkatesan B, Choudhury GG. Mesangial cell hypertrophy by high glucose is mediated by downregulation of the tumor suppressor PTEN. Diabetes 55:2115–2125. 2006;


151. Wu D, Peng F, Zhang B, et al. PKC-β1 mediates glucose-induced Akt activation and TGF-β1 upregulation in mesangial cells. J Am Soc Nephrol 20:554–566. 2009;



152. Xin X, Khan ZA, Chen S, Chakrabarti S. Glucose-induced Akt1 activation mediates fibronectin synthesis in endothelial cells. Diabetologia 48:2428–2436. 2005;


153. Shemesh II, Rozen-Zvi B, Kalechman Y, Gafter U, Sredni B. AS101 prevents diabetic nephropathy progression and mesangial cell dysfunction: regulation of the AKT downstream pathway. PLoS One 9:e1142872014;



154. Sun L, Zhang D, Liu F, et al. Low-dose paclitaxel ameliorates fibrosis in the remnant kidney model by down-regulating miR-192. J Pathol 225:364–377. 2011;



155. Pan Y, Jia T, Zhang Y, et al. MS2 VLP-based delivery of microRNA-146a inhibits autoantibody production in lupusprone mice. Int J Nanomedicine 7:5957–5967. 2012;



156. Woo JS, Kim VN. MeCP2 caught moonlighting as a suppressor of microRNA processing. Dev Cell 28:477–478. 2014;


157. Bracaglia G, Conca B, Bergo A, et al. Methyl–CpG–binding protein 2 is phosphorylated by homeodomain–interacting protein kinase 2 and contributes to apoptosis. EMBO Rep 10:1327–1333. 2009;



158. Liu R, Das B, Xiao W, et al. A novel inhibitor of homeodomain interacting protein kinase 2 mitigates kidney fibrosis through inhibition of the TGF-β1/Smad3 pathway. J Am Soc Nephrol 28:2133–2143. 2017;



159. Roos M, Pradère U, Ngondo RP, et al. A small-molecule inhibitor of Lin28. ACS Chem Biol 11:2773–2781. 2016;


160. Lightfoot HL, Miska EA, Balasubramanian S. Identification of small molecule inhibitors of the Lin28-mediated blockage of pre-let-7g processing. Org Biomol Chem 14:10208–10216. 2016;



161. Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 3462014.

162. Dominguez AA, Lim WA, Qi LS. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol 17:5–15. 2016;


163. Liao HK, Hatanaka F, Araoka T, et al. In vivo target gene activation via CRISPR/Cas9-mediated trans-epigenetic modulation. Cell 171:1495–1507.e15. 2017;



164. Komor AC, Badran AH, Liu DR. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168:20–36. 2017;


165. Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533:420–424. 2016;



166. Nishida K, Arazoe T, Yachie N, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353:aaf87292016;


167. Kim YB, Komor AC, Levy JM, Packer MS, Zhao KT, Liu DR. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol 35:371–376. 2017;



168. Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551:464–471. 2017;



169. Mali P, Aach J, Stranges PB, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 31:833–838. 2013;



170. Perez-Pinera P, Kocak DD, Vockley CM, et al. RNA-guided gene activation by CRISPR-Cas9–based transcription factors. Nat Methods 10:973–976. 2013;



171. Gilbert LA, Horlbeck MA, Adamson B, et al. Genomescale CRISPR-mediated control of gene repression and activation. Cell 159:647–661. 2014;



172. Kearns NA, Pham H, Tabak B, et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods 12:401–403. 2015;



173. Liu XS, Wu H, Ji X, et al. Editing DNA methylation in the mammalian genome. Cell 167:233–247.e217. 2016;



174. Hilton IB, D’Ippolito AM, Vockley CM, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33:510–517. 2015;






 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print